Research Interests:
 Prediction of molecular and spectroscopic
properties (UV, Infrared, Raman and resonance Raman) and reaction pathways with
a special focus on transition and heavy metals compounds. Various applications
of electronic structure calculation to enhance understanding of the structure
and function of native metals in proteins and toxicity of heavy metals.
Research interests and computational application span wide areas of modern
chemistry, biochemistry, geochemistry and environmental science.
Prediction of molecular and spectroscopic
properties (UV, Infrared, Raman and resonance Raman) and reaction pathways with
a special focus on transition and heavy metals compounds. Various applications
of electronic structure calculation to enhance understanding of the structure
and function of native metals in proteins and toxicity of heavy metals.
Research interests and computational application span wide areas of modern
chemistry, biochemistry, geochemistry and environmental science.
Read a short article in “Brooklyn
College Magazine” Fall 2006 issue, page 9 about Jarzecki’s research
interests.
Computers
can help to understand the role of metals in biology:
 From all elements of the periodic table, carbon,
hydrogen, nitrogen, oxygen, phosphorus, and sulfur are the big six building
blocks which found the way to all chemistry and biochemistry textbooks as major
cellular components of life such as proteins, nucleic acids, lipids-membranes,
polysaccharides, and metabolites. Nevertheless it is now known that life cannot
survive with only these principle elements and at least 20 additional
“inorganic” elements are essential for most forms of life, yet some elements such
as lead, arsenic and mercury are extremely poisonous. Through countless years
of evolution nature has adopted these essential elements of life to perform
diverse functions and processes which include signaling, charge balance and
transfer, energy transport and storage, bio mineralization and many others.
Toxic elements usually follow the life paths of essential elements but by
introducing new coordination preferences and new chemistry they interrupt or
inhibit life supporting processes.
From all elements of the periodic table, carbon,
hydrogen, nitrogen, oxygen, phosphorus, and sulfur are the big six building
blocks which found the way to all chemistry and biochemistry textbooks as major
cellular components of life such as proteins, nucleic acids, lipids-membranes,
polysaccharides, and metabolites. Nevertheless it is now known that life cannot
survive with only these principle elements and at least 20 additional
“inorganic” elements are essential for most forms of life, yet some elements such
as lead, arsenic and mercury are extremely poisonous. Through countless years
of evolution nature has adopted these essential elements of life to perform
diverse functions and processes which include signaling, charge balance and
transfer, energy transport and storage, bio mineralization and many others.
Toxic elements usually follow the life paths of essential elements but by
introducing new coordination preferences and new chemistry they interrupt or
inhibit life supporting processes.
 Recent advances in biological inorganic chemistry
have already impacted both environmental science and medicine but still many
relationships between structure and functions remain unknown and new
fascinating relationships are being discovered. Modern experimental techniques
are developed to understand, elucidate and to reveal structural, mechanistic
and genetic underpinnings of these relationships. With the recent great
advances in computer technologies, for the first time the state-of-the-art
methods of quantum chemistry might have a significant role in design,
elucidation, interpretation and guiding
these novel experiments and measurements.
Recent advances in biological inorganic chemistry
have already impacted both environmental science and medicine but still many
relationships between structure and functions remain unknown and new
fascinating relationships are being discovered. Modern experimental techniques
are developed to understand, elucidate and to reveal structural, mechanistic
and genetic underpinnings of these relationships. With the recent great
advances in computer technologies, for the first time the state-of-the-art
methods of quantum chemistry might have a significant role in design,
elucidation, interpretation and guiding
these novel experiments and measurements.
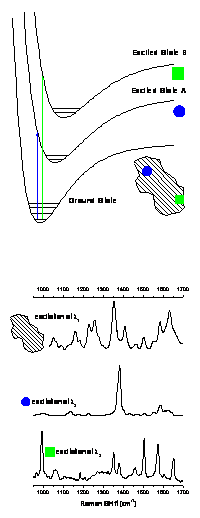 Computational modeling in Jarzecki’s research
laboratory focus on structure, function, spectroscopic properties and
biological paths of the “life” elements, and their changes caused by the toxic
elements. With the financial support from the National Institutes of Health he
initiated a systematic computational methodology aimed at elucidating the
essential connections between lead coordination preferences and lead
toxicity. He has developed a reliable
modeling of resonance Raman intensity patterns emerging from the UV excitations
of lead-thiolate charge transfer bands commonly observed in lead substituted
proteins. These simulations allow for spectroscopic characterization of various
lead sites in proteins and to bridge spectroscopic observations with the
structure and function of lead-binding sites leading to understanding lead
poisoning at the molecular level. Jarzecki’s work is designed to exploit the
power of computational modeling, in combination with resonance Raman
spectroscopy and to provide a fruitful interaction between computations and
experiment. Application of similar computational strategies to model other
heavy metal toxic ions such as mercury, arsenic, cadmium and chromium are also
in progress.
Computational modeling in Jarzecki’s research
laboratory focus on structure, function, spectroscopic properties and
biological paths of the “life” elements, and their changes caused by the toxic
elements. With the financial support from the National Institutes of Health he
initiated a systematic computational methodology aimed at elucidating the
essential connections between lead coordination preferences and lead
toxicity. He has developed a reliable
modeling of resonance Raman intensity patterns emerging from the UV excitations
of lead-thiolate charge transfer bands commonly observed in lead substituted
proteins. These simulations allow for spectroscopic characterization of various
lead sites in proteins and to bridge spectroscopic observations with the
structure and function of lead-binding sites leading to understanding lead
poisoning at the molecular level. Jarzecki’s work is designed to exploit the
power of computational modeling, in combination with resonance Raman
spectroscopy and to provide a fruitful interaction between computations and
experiment. Application of similar computational strategies to model other
heavy metal toxic ions such as mercury, arsenic, cadmium and chromium are also
in progress.
